1. About CMS 2.0¶
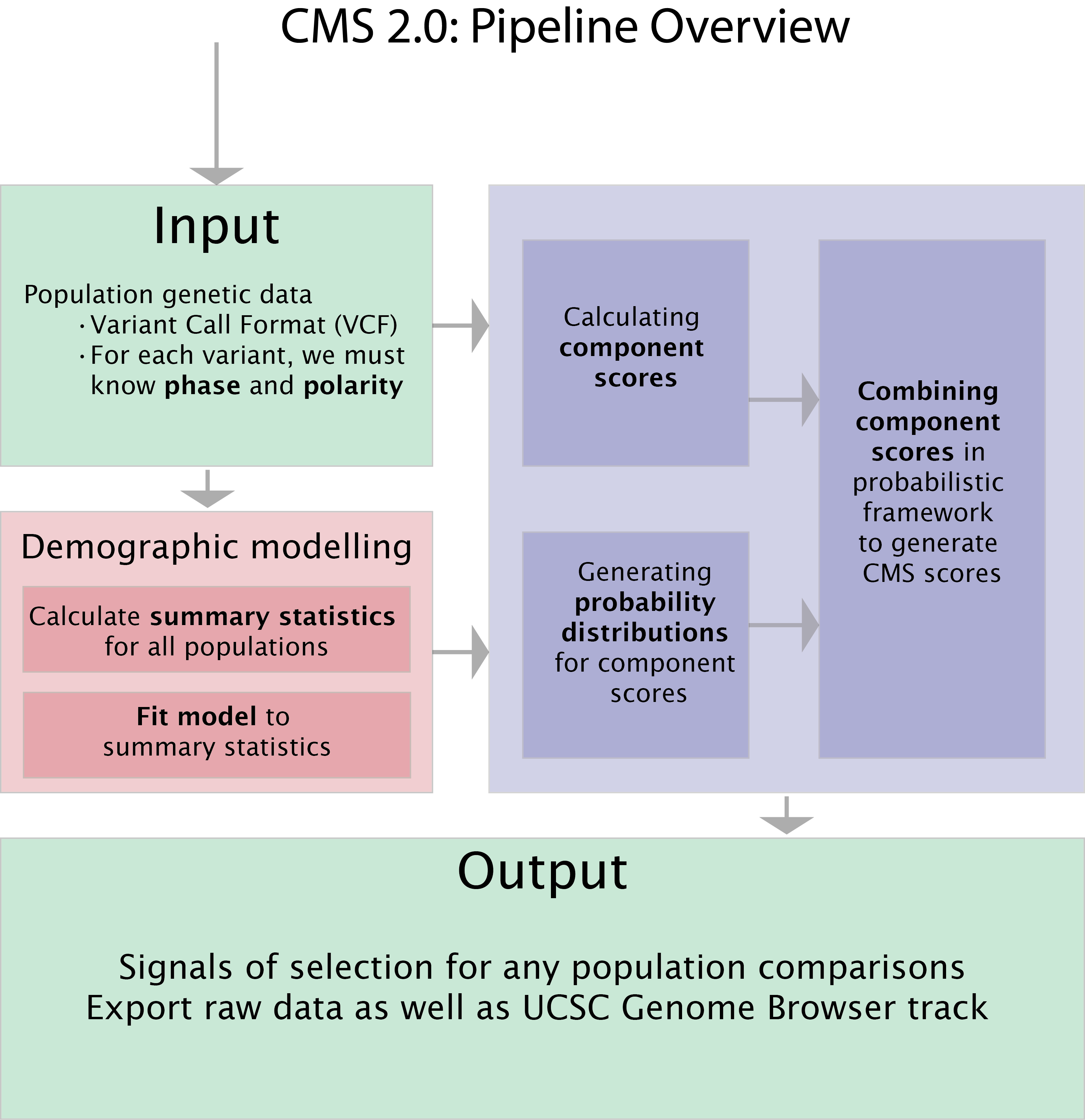
Composite of Multiple Signals (CMS) refers to a family of tests applied to population genetic datasets in order to (i) identify genomic regions that may have been subject to strong recent positive selection (a ‘sweep’) and (ii) to narrow signals of selection within such regions, in order to identify tractable lists of candidate variants for experimental scrutiny. In both of these cases, CMS requires (a) phased variation data for several populations, along with (b) the identity of the ancestral allele for a majority of sites listed. It was developed with humans in mind (e.g., the 1000 Genomes Project) but could in principle be applied to any diploid species with data in VCF or TPED format.
In its current instantiation (‘CMS 2.0’), it includes scripts to (i) calculate a variety of selection metrics for each population, (ii) model the demographic history of the dataset using an exploratory approach, (iii) generate probability distributions for each selection metric from data simulated from demographic models, (iv) generate composite scores and (v) visualize signals of selection in the UCSC Genome Browser.
1.1. Background¶
The method used in CMS is described in greater detail in the following papers:
A Composite of Multiple Signals distinguishes causal variants in regions of positive selection Sharon R. Grossman, Ilya Shylakhter, Elinor K. Karlsson, Elizabeth H. Byrne, Shannon Morales, Gabriel Frieden, Elizabeth Hostetter, Elaine Angelino, Manuel Garber, Or Zuk, Eric S. Lander, Stephen F. Schaffner, and Pardis C. Sabeti Science 12 February 2010: 327 (5967), 883-886.Published online 7 January 2010 [DOI:10.1126/science.1183863]
Identifying recent adaptations in large-scale genomic data Grossman SR, Andersen KG, Shlyakhter I, Tabrizi S, Winnicki S, Yen A, Park DJ, Griesemer D, Karlsson EK, Wong SH, Cabili M, Adegbola RA, Bamezai RN, Hill AV, Vannberg FO, Rinn JL; 1000 Genomes Project, Lander ES, Schaffner SF, Sabeti PC. Cell 14 February 2013: 152 (4), 883-886.Published online 7 January 2010 [DOI:10.1016/j.cell.2013.01.035]
1.2. Coalescent simulations¶
CMS uses simulated population genetic data for a variety of purposes. For the purpose of flexibility, this pipeline is optimized for use with cosi 2, but it would theoretically be straightforward to substitute other simulators supporting selection.
Cosi2: an efficient simulator of exact and approximate coalescent with selection. Shlyakhter I, Sabeti PC, Schaffner SF. Bioinformatics 1 December 2014: 30 (23), 3427-9.Published online 22 August 2014 [DOI:10.1093/bioinformatics/btu562]